Affinity-maturation DEL technology
While most single-pharmacophore DELs described so far feature DNA in double-stranded format, we used single-stranded DNA barcodes in order to enable a modular lead expansion procedure by DNA self-assembly.
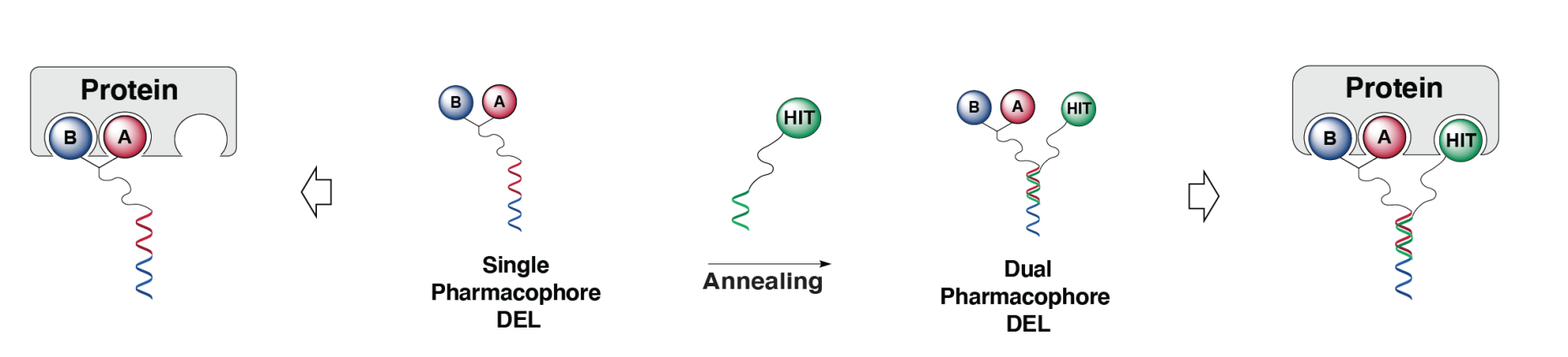
A novel, versatile DEL was constructed based on a combination of a two-building block single-pharmacophore DEL with a known hit on the pairing strand (yielding a dual-display) [1].
Two sets of building blocks were coupled to a central stereo-defined 4-azido-5-methoxy-5-oxopentanoic acid. The resulting single-pharmacophore GB-DEL library was screened as such against multiple targets, allowing the discovery of novel binders which were compatible with Lipinksi’s rule of five.
Using the modular dual-display, novel protein ligands could efficiently be discovered from the dual-pharmacophore affinity maturation libraries. This approach termed "2+1" ESAC" is an important advancement of the ESAC concept that we had previously proposed [2],[3] and allowed us to affinity-mature known protein ligands of low to medium affinity which, in turn, may be used in an iterative fashion [1].
[1] Bassi, G., Favalli, N., Vuk, M., Catalano, M., Martinelli, A., Trenner, A., Porro, A., Yang, S., Tham, C. L., Moroglu, M., Yue, W. W., Conway, S. J., Vogt, P. K., Sartori, A. A., Scheuermann, J.* & Neri, D.* (2020) Adv. Sci. 7, 2001970.
external page https://doi.org/10.1002/advs.202001970
[2] Melkko, S.*, Scheuermann, J.*, Dumelin, C. E., Neri, D. (2004),
Nat. Biotechnol. 22, 568-74
external page https://doi.org/10.1038/nbt961
[3] Wichert, M., Krall, N., Decurtins, W., Franzini, R. M., Pretto, F., Schneider, P., Neri, D., Scheuermann, J. (2015),
Nat. Chem. 7, 241-9.
external page https://doi.org/10.1038/nchem.2158