Dual-display DEL technology
Dual-pharmacophore libraries are often constructed on the basis of a synthetic strategy, leading to “encoded self-assembled chemical (ESAC) libraries”. This technology, initially proposed by our group in 2004 [1], makes use of two sets of partially complementary oligonucleotides, each containing a distinctive DNA barcode (see Figure, left). The two sets of chemically modified oligonucleotides constitute “sub-libraries”, which can be mixed and hybridized to form stable DNA-heteroduplexes, which display pairs of building blocks. For example, the combinatorial assembly of two sub-libraries with 1’000 chemical moieties each leads to a 1-million membered ESAC library. In order to generate ESAC libraries with DNA structures, which allow the identification of the chemical nature of the pairs of building blocks by DNA sequencing, an encoding strategy featuring the use of oligonucleotides with abasic sites has been described (Figure 9, left). This feature allows the simultaneous determination of the binding fragments by PCR amplification and high-throughput DNA sequencing [2].
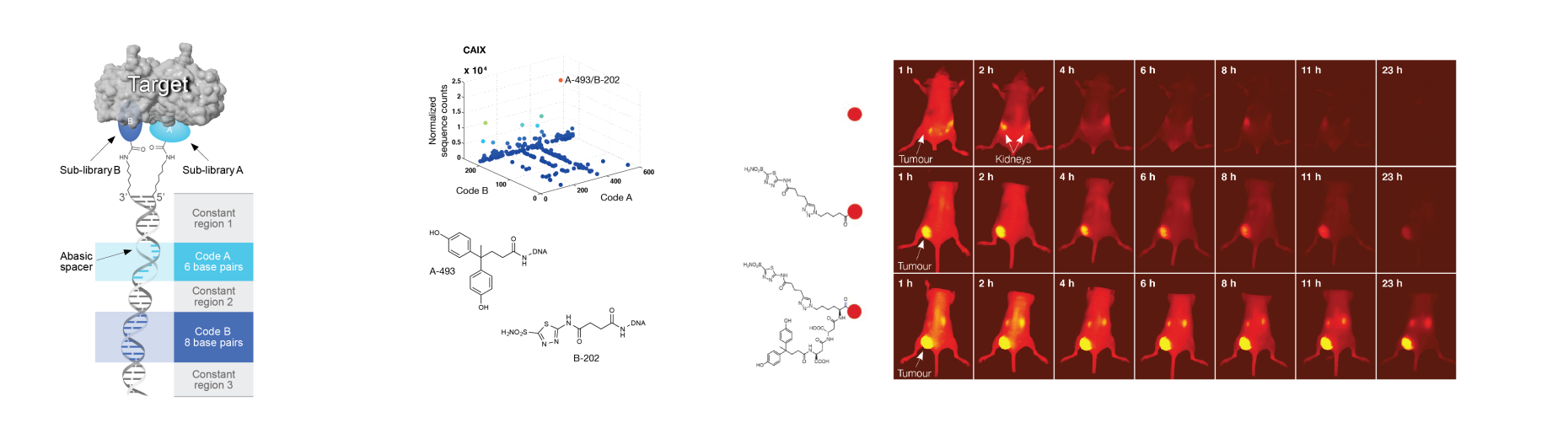
Dual-display libraries feature a higher degree of flexibility in the arrangement of the chemical building blocks compared to single-pharmacophore DECLs and may more easily reach adjacent binding sites on the surface of the cognate protein target. On the other hand, as soon as a combination of building blocks (A and B) has been identified as preferential binding pair, various linkers need to be tested, in order to yield binding molecules devoid of DNA, which can be used for practical applications. For that purpose, the use of a set of predefined bifunctional scaffolds and the implementation of affinity measurements on locked-nucleic acids (LNA) [3] may facilitate the identification of the best linker [4].
Dual-pharmacophore libraries based on ESAC technology have yielded an acetazolamide derivative (Figure above, right, 3rd row) that bound to carbonic anhydrase IX with a dissociation constant in the sub-nanomolar range [2]. The same ESAC library also produced a 190nM binder against alpha-1-acid glycoprotein (AGP), thanks to the synergistic action of two building blocks that mediated a potent chelate binding to the target protein. Interestingly, the individual building blocks did not display any detectable protein binding when used on their own. The optimization of the linker connecting the two building blocks led to an improved binder with a dissociation constant of 9.9nM [4].
[1] Melkko, S.*, Scheuermann, J.*, Dumelin, C. E., Neri, D. (2004),
Nat. Biotechnol. 22, 568-74
external page https://doi.org/10.1038/nbt961
[2] Wichert, M., Krall, N., Decurtins, W., Franzini, R. M., Pretto, F., Schneider, P., Neri, D., Scheuermann, J. (2015),
Nat. Chem. 7, 241-9.
external page https://doi.org/10.1038/nchem.2158
[3] Zimmermann, G., Li, Y., Rieder, U., Mattarella, M., Neri, D., Scheuermann, J. (2017),
ChemBioChem. 18, 853-857.
external page https://doi.org/10.1002/cbic.201600637
[4] Bigatti, M., Dal Corso, A., Vanetti, S., Cazzamalli, S., Rieder, U., Scheuermann, J., Neri, D., Sladojevich, F. (2017), ChemMedChem. 12, 1748-1752.
external page https://doi.org/10.1002/cmdc.201700569