DNA-Encoded Chemical libraries (DEL)
Traditionally, the search for small molecule binders for a protein of pharmaceutical interest has been performed by testing individual molecules one by one for their ability to bind to a particular protein of interest. This process (“high-throughput screening”) is very expensive and requires complex logistics. Even the largest pharmaceutical companies cannot generate and screen compound collections (“libraries”) larger than a few million compounds. As a result, screening campaigns of conventional libraries often fail to deliver binders (“hits”) against proteins of interest.
DNA-encoded chemical libraries (DELs) are collections of organic molecules, individually tagged with DNA fragments serving as amplifiable identification barcodes. DEL technology allows the construction and screening of compound collections of unprecedented size (e.g., comprising billions of compounds) and chemical diversity, thus representing a revolution in the field of small-molecule drug discovery:[1] the basic principle of linking phenotype and genotype for selection can also be applied to the “world” of small organic molecules. Over the last 20 years this has led to the now industrially established DEL technology. This encoding process was first postulated in a theoretical paper by Sidney Brenner and Richard Lerner in 1992.[2]
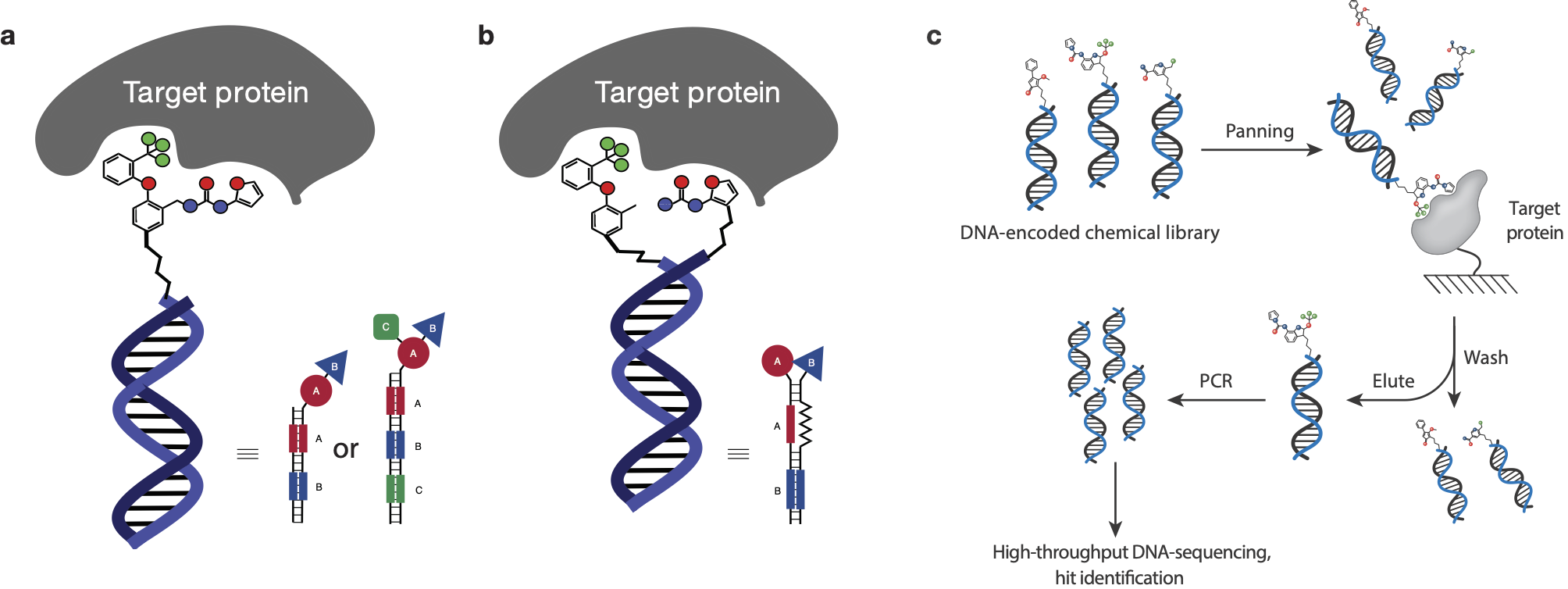
DELs are collections of organic molecules, individually tagged with DNA fragments serving as amplifiable identification barcodes [1]. Molecules can be displayed on one DNA strand ("single-pharmacophore DELs", Figure 1a) or on both DNA strands forming a DNA heteroduplex ("dual-pharmacophore DELs, Figure 1b). These DELs can contain billions and more individual chemical entities in a small container (e.g., an Eppendorf tube).
Due to the ultrasensitive DNA readout by polymerase chain reaction (PCR) DEL technology allows the screening of compound collections of unprecedented size at once (e.g., comprising billions of compounds): DELs can be subjected to affinity-based selections on target antigens of interest and binders that are retained on the resin after washing can be identified after PCR and high-throughput DNA sequencing by comparing the relative composition of the library (i.e., the frequency of all compounds in the library) before and after the selection experiment (Figure 1c).[3] Using dedicated software tools, the “counts” and thus the enrichment of individual library members can be displayed in dedicated graphs.[3]
Dario Neri (now CEO of Philogen Ltd.) and Jörg Scheuermann have pioneered DEL technology at ETH since 2002 and hold the track record of >60 publications in this growing research field.
[1] a) Favalli N, Bassi G, Scheuermann J, Neri D. (2018) FEBS Lett. 592:2168–2180
external page https://doi.org/10.1002/1873-3468.13068
[1] b) Neri D, Lerner RA. (2018) Annu Rev Biochem. 87:479–502
external page https://doi.org/10.1146/annurev-biochem-062917-012550
[1] c) Goodnow RA, Dumelin CE, Keefe AD. (2017) Nat Rev Drug Discov. 16:131–147
external page https://doi.org/10.1038/nrd.2016.213
[1] d) Franzini RM, Neri D, Scheuermann J. (2014). Acc Chem Res. 47:1247–1255
external page https://doi.org/10.1021/ar400284t
[2] Brenner S, Lerner RA. (1992) Proc. Natl Acad. Sci. USA 89, 5381–5383
external page https://doi.org/10.1073/pnas.89.12.5381
[3] Decurtins W, Wichert M, Franzini RM, Buller F, Stravs MA, Zhang Y, Neri D., Scheuermann, J (2016). Nature protocols, 11(4), 764–780.
external page https://doi.org/10.1038/nprot.2016.039